This feature—expanding on a previous data use story—is also published in the GBIF Science Review 2019, which highlights important and noteworthy examples of the use and reuse of GBIF-mediated data in research and policy.
In early May of 2018, Texas State University (TSU) alumnus Mark Deka submitted a study modelling Nipah virus (NiV) infection risk in South and Southeast Asia for publication in the journal Tropical Medicine and Infectious Disease. Part of his PhD, the paper had been in the works for nearly a year and a half.
Working in the area of medical geography, Mark—originally from Cleveland, Ohio—was interested in vector-borne diseases and the geospatial and geographic components of disease transmission.
“A lot of people don’t realize the role of geography and spatial gradients in relation to distribution of communicable diseases for hosts and susceptible populations”, says Deka, who came to Texas in 2015 and started his career working on tick-borne diseases. It was during this time he started looking into using GBIF-mediated data for creating models for his research. This proved useful when he later turned to NiV research.
A relative of the Hendra virus that takes its name from the Malaysian village in which it was first isolated from pigs, the NiV causes encephalitis in infected organisms. Flying foxes (Pteropus sp.) are the presumed wildlife reservoir of NiV, and consumption of raw date palm (Phoenix dactylifera) sap contaminated with Pteropus urine or saliva is believed to be an important risk factor.
Mark’s study combined biosurveillance data with GBIF-mediated Pteropus occurrences to create models mapping infection risk hot spots in South and Southeast Asia while identifying environmental features associated with increased risk.
“The data from GBIF.org played a huge role. While some people just build a single model to show human transmission cases, we were interested in mapping the overlap between several models and also measuring it. The occurrence data for the flying foxes was very important, and there was a lot of it, too!”
The results of the study showed high risk of disease in a combined area of near 3 million sq. km, or 19 per cent of the total study area—including several risk hot spots in South India, in which no outbreak had previous occurred.
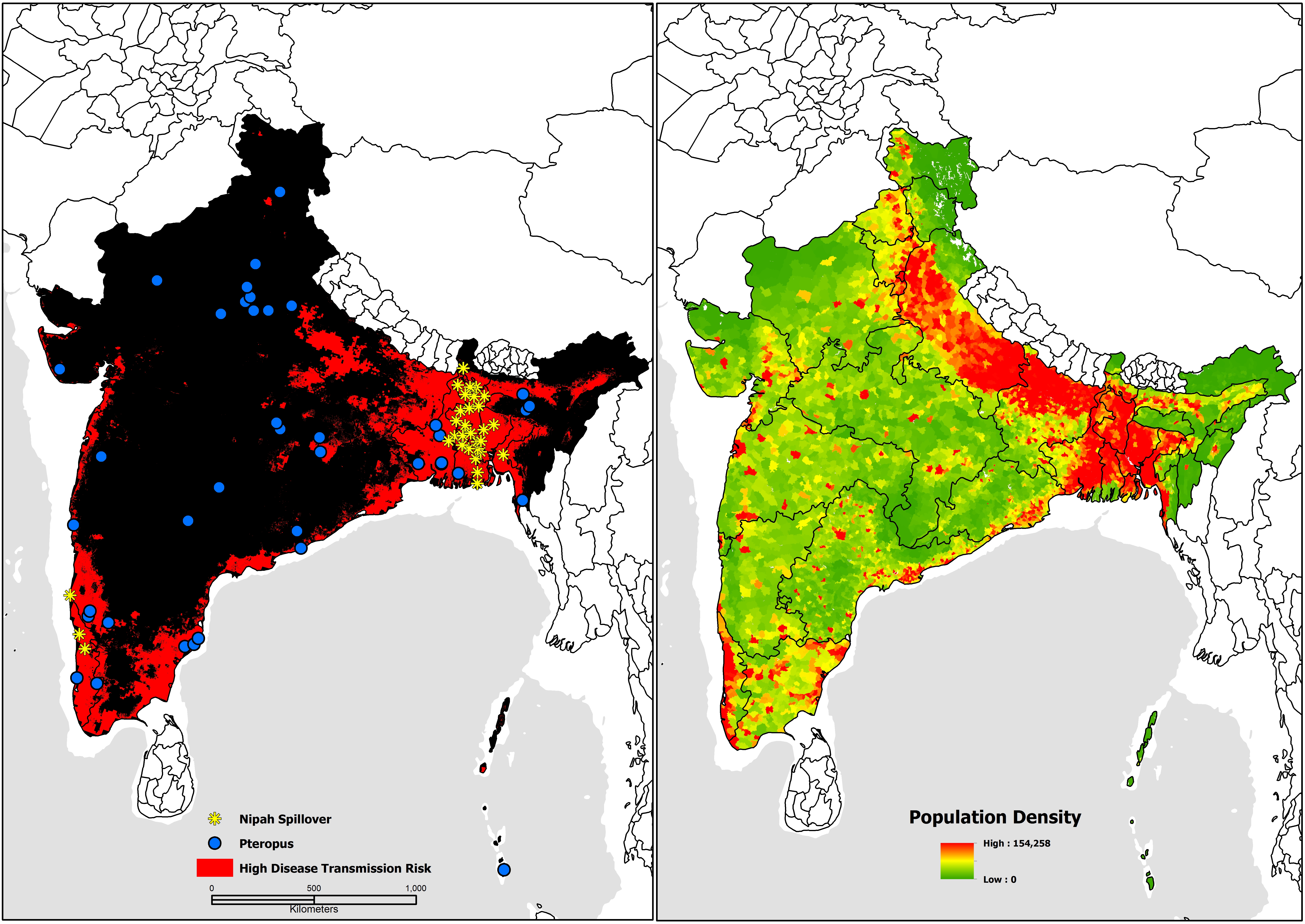
Nipah virus disease transmission risk in Bangladesh and India with updated human case locations from May 2018. Figure from Deka M and Morshed N (2018) (CC BY 4.0)
On 19 May 2018—within weeks of Mark and colleagues submitting their paper—the Indian Government’s National Centre for Disease Control and the World Health Organization reported a NiV disease outbreak from Kozhikode district of Kerala, India—the first in South India. Within less than two weeks 18 people had contracted the disease and 17 were dead.
“We were in the process of submitting the final edits when the outbreak in Kerala took place. On the one hand, it’s very exciting to see that your models can make accurate predictions, but knowing the consequences of these outbreaks puts an immediate dampener on your excitement.”
Since the study was published, Mark was approached by a vaccine consultant in Germany who wanted to understand if the results of the study could help them in their work.
“They’re trying to develop a vaccine, figure out how many vaccines to develop and who to target. They were talking about vaccine stockpiles, but I had to explain that I am a geographer. I know nothing about vaccine stockpiles! I told them to target the people at highest risk—the people who work in date palm cultivation. If you’re going to do a vaccine, target them first.”
Having completed his PhD at TSU, Mark is now working for the Centers for Disease Control and Prevention (CDC) in Atlanta, Georgia, where he will be working on similar medical geography modelling with a focus on bacterial diseases.